Background
β-Galactosidase is encoded by the lacZ gene of the lac operon in E. coli. It is a large (120 kDa, 1024 amino acids) protein that forms a tetramer. The enzyme’s function in the cell is to cleave lactose to glucose and galactose so that they can be used as carbon/energy sources. The synthetic compound o-nitrophenyl-β-D-galactoside (ONPG) is also recognized as a substrate and cleaved to yield galactose and o-nitrophenol which has a yellow color. When ONPG is in excess over the enzyme in a reaction, the production of o-nitrophenol per unit time is proportional to the concentration of β-Galactosidase; thus, the production of yellow color can be used to determine enzyme concentration.
So, why do we care? Usually, experiments are designed so that the β-Galactosidase concentration in the cell is a readout for some aspect of a system being studied. For example, an investigator may fuse a promoter to the lacZ gene and use β-Gal levels as a readout for promoter activity under various conditions. In 1972, Jeffrey Miller published “Experiments in Molecular Genetics” which contained a protocol for determining the amount of β-Gal with ONPG. Because of this, ONPG/β-Gal assays are referred to as “Miller” assays, and a standardized amount of β-Gal activity is a “Miller Unit”.
1 Miller Unit = [math]\displaystyle{ 1000 * \frac{(Abs_{420} – (1.75*Abs_{550}))}{(t * v * Abs_{600})} }[/math]
where:
- Abs420 is the absorbance of the yellow o-nitrophenol,
- Abs550 is the scatter from cell debris, which, when multiplied by 1.75 approximates the scatter observed at 420nm,
- t = reaction time in minutes,
- v = volume of culture assayed in milliliters,
- Abs600† reflects cell density.
†Note that this value is different for each spectrophotometer used and should be calibrated by plating dilutions of known Abs600 cultures to determine the colony-forming units per Abs600.
In his book, Dr. Miller explains that this formula yields approximately 1 Miller Unit for uninduced E. coli (low β-Gal production) and approximately 1000 units for a fully induced culture (grown on lactose or IPTG).
In my experience, cultures of MG1655 induced with 1 mM IPTG in log phase have 1500-1800 Miller units. The reason for the difference is not known, but I suspect it stems from differences in the Abs600/cell density between Dr. Miller’s spectrophotometer and the one I use and the fact I do my Miller assays at 30 °C (for convenience) whereas Dr. Miller performed his assays at 28 °C. I have made promoter fusions that generate ~40,000 Miller units; however, as will be discussed below, this is too high for the assay and so the protocol was changed to lower this value.
Protocol
The protocol I use is derived from a paper by Zhang and Bremer (JBC 270, 1995, Free full text!) in which the original Miller protocol was greatly simplified to allow more samples to be measured with less manipulation.
In short, the protocol consists of measuring the cell density of a culture of bacteria (Abs600), then removing an aliquot of the cells from the cuvette and mixing them with a “permeabilization” solution that contains detergent which disrupts the cell membranes (but leaves the β-Gal intact). This kills the cells and stops translation. After incubation, an ONPG “substrate” solution is added and the yellow color allowed to develop. A “stop” solution is then added and the absorbance of o-nitrophenol is measured.
- Grow cultures under whatever conditions you wish to test.
- During growth, pre-measure 80 μL aliquots of permeabilization solution into 1.5 mL microfuge tubes and close them.
- Measure Abs600 and RECORD IT!
- Remove a 20 μL aliquot of the culture and add it to the 80 μL of permeabilization solution.
The sample is now stable for several hours. This allows you to perform time-course experiments.
- After the last sample is taken, move the samples and the Substrate solution to the 30 °C warm room for 20-30 minutes.
- Add 600 μL of Substrate solution to each tube and NOTE THE TIME OF ADDITION.
- After sufficient color has developed, add 700 μL of Stop solution, mix well, and NOTE THE STOP TIME.
- After stopping the last sample (some may take longer than others, but generally they are done in 30-90 minutes), transfer the tubes to a microfuge and spin for 5-10 minutes at full speed.
- Carefully remove the tubes from the centrifuge and transfer solution from the TOP of the tubes to your cuvette(s). You are trying to avoid having particulate material in the cuvette so that scattering will not influence the reading.
- Record Abs420. This should be less than 1 and greater than 0.05. If it’s a bit outside of this range, don’t sweat it.
Calculate Miller Units as:
[math]\displaystyle{ 1000 * \frac{(Abs_{420})}{((Abs_{600} \text{ of culture sampled})*(\text{volume } [0.02 \text{ mL}])*(\text{reaction time}))} }[/math]
Comments on the assay
- Reshma 11:28, 15 October 2007 (CDT): Miller recommends a culture with OD600 = 0.28 to 0.70. However, he claims that overnight cultures can also be used but that exponentially growing cells give more precise assays.
- When is the reaction done? I never found a good answer for this in the literature. If I let a reaction go to completion, I measured an Abs420 of ~2-3. Of course, this is out of the reliable range of the spectrophotometer, but it gives an indication of how far the reaction can go. I got reproducible data when the yellow color was just detectable before adding the stop solution up to about the color of LB broth before stopping. Remember, you need the substrate to saturate the enzyme during the course of the reaction, so don’t let them go too far. I routinely make three separate measurements for each culture and average them.
- Reshma 11:28, 15 October 2007 (CDT): Miller recommends that the OD420nm reading should ideally be 0.6-0.9.
- Frequently, people use disposable plastic cuvettes to measure both the culture turbidity and the yellow o-nitropenol. It is my experience that the disposable cuvetees have HORRIBLE optics: yes, they are clear, but the light path is pathetically distorted (just look through one at a distant object). This is not a big problem if the same cuvette is used for the blank and the sample and it is oriented in the same way in the spectrophotometer. The problem arises when many different samples are measured in different cuvettes. The values can can vary GREATLY even for the same culture measured in different cuvettes. I recommend using either a high quality glass or quartz cuvette, or measuring the samples in a plate reader using flat-bottomed 96 well plates. I have found my error in pipetting 150 μL of culture for Abs600 measurements was WAY less than using disposable cuvettes. Of course, the turbidity measured should be calibrated to cells/mL as with a 1cm cuvette.
- By spinning the samples and carefully removing the a sample from the supernatant, you avoid having to measure the scattering at 550nm and guessing what it would be at 420nm.
- If your reaction has too much β-Gal, the tube will turn yellow in a few minutes (or even seconds). This is too fast. One of the greatest contributions to error will be your estimate of reaction time. By having the reactions conditions set so that it takes about an hour, the time errors become insignificant. If you need to slow the reaction, you can use fewer cells and increase the amount of permeabilization buffer so the volume is still 100 μL. Alternately, you can re-engineer the ribosome-binding site of your β-Gal construct to weaken it. I found that if my cells were making 40,000 Miller units of β-Gal, they were very sick from the translation stress. It was better in this case to weaken the translation of the β-Gal mRNA.
- I do these reactions like I do enzyme kinetics. I start the samples 10 sec apart (with the time counting up) so it is possible to get accurate reaction times even if you only let your reactions go for a few minutes. I get very reproducible results with reaction times of 1.5-30 min. Once you get a feel for how long your reactions will need, it is easy to group samples that will need the same reaction time. Doing each sample in triplicate will give you some confidence that your timing is reproducible.–Kathleen 16:43, 14 December 2005 (EST)
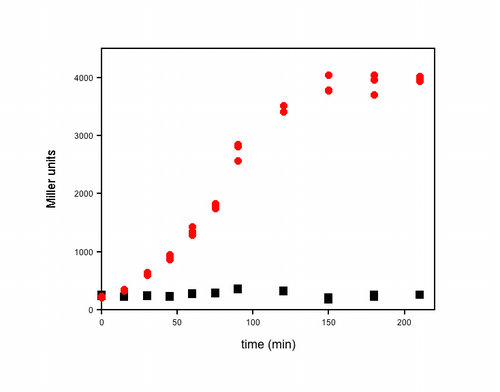
- Here’s an example of some actual data I obtained using this assay. It was a timecourse experiment. At each time, I removed 1 mL from each of my cultures, measured the OD600, took three 20 µL aliquots directly from the cuvette and added each to 80 µL of permeabilization solution. I performed the assay exactly as described above, and all of the samples were kept at room temperature until the timecourse was finished. The OD600 for these cultures varied from 0.4 to 4 (in my spec) over the course of this experiment and the reaction times for the β-Gal assays varied from 2–25 min. The three individual β-Gal assays for each time point for each culture (red or black symbols) are plotted in the graph to illustrate the reproducibility of the assay within each experiment.–Kathleen
Recipes
Permeabilization Solution
You need 80 μL per sample.
- 100 mM dibasic sodium phosphate (Na2HPO4)
- (The Zhang protocol has 200 mM sodium phosphate. I could never get this into solution with the other components, no matter what I tried so I backed it off to 100 mM. I have even used 50 mM with no detectable change.)
- 20 mM KCl
- 2 mM MgSO4
- 0.8 mg/mL CTAB (hexadecyltrimethylammonium bromide)
- 0.4 mg/mL sodium deoxycholate
- 5.4 μL/mL beta-mercaptoethanol
Substrate solution
You need 600 μL per sample.
- 60 mM Na2HPO4
- 40 mM NaH2PO4
- 1 mg/mL o-nitrophenyl-β-D-Galactoside (ONPG)
- 2.7 μL/mL β-mercaptoethanol
(The Zhang protocol also has 20 μg/mL CTAB and 10 μg/mL deoxycholate. I leave these out figuring that there is still plenty from the permeabilization solution and, if they ain’t dead yet, they ain’t gonna be.)
Stop solution
You need 700 μL per sample.
- 1 M Sodium Carbonate (Na2CO3)
The high pH of the stop solution denatures the β-Gal and approximately doubles the yellow color of the reaction.