Materials
- For making Spots:
- 1% Cresol Red
- DNA (100 ng/μL)
- Crane & Co. 100% cotton, acid free thesis paper
- For using spots to transform:
- Harris Uni-Core Punch, 2mm and Olfa Cutting Mat
- TE
- Competent Cells
Procedure
Making Spots
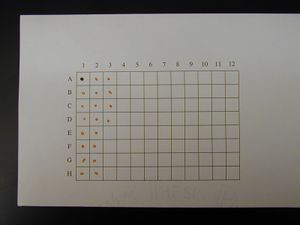
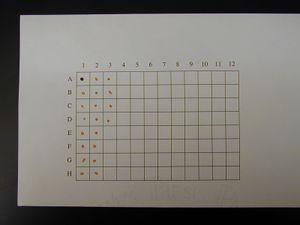 Spotted Grid
Spotted Grid
- Mix 1 part 1% Cresol Red with 4 parts DNA that is at least 100 ng/μL
- The exact amounts depend on how many 2 μL spots you plan to make
- Using the excess of Cresol Red is helpful for when you transform, since you can visibly tell which cells have had DNA added and which have not
- Make 2 μL spots on 100% cotton, acid free thesis paper
- Place a second sheet of paper under the one to be spotted to keep it free from contamination
- Leave spots to dry at room temperature. This takes between 45 minutes and an hour
- Once dry, spots can be used right away or stored at -20 °C. To store – place the spotted paper between two others to protect it.
Using spots to transform E. coli
 Olfa cutting mat and Uni-Core Punch
Olfa cutting mat and Uni-Core Punch
 Punched Spot
Punched Spot
- Cut out spot from surrounding paper using the Uni-Core Punch on the Olfa cutting mat
- Soak spot in 20 μL TE for 15 minutes.
- Thaw competent cells on ice while spots soak
- I used TOP10 chemically competent cells
- Add 5 μL of the TE the spot soaked in to 50 μL competent cells
- Spots could also be directly added to cells. Soaking in TE is better though, since there is DNA left over if the transformation does not work for some reason.
- Incubate cells on ice for 30 minutes
- Heat shock cells at 43 °C
- If cells are in individual tubes, heat shock for 30 seconds. If cells are in a 96-well plate extend the heat shock to 1 minute.
- Incubate cells on ice for 2 minutes
- Add 200 μL SOC
- Incubate at 37 °C for 2 hours
- Spread cells on previously made LB plates with proper antibiotic
- Grow overnight at 37 °C