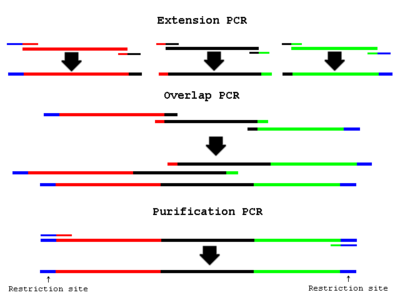
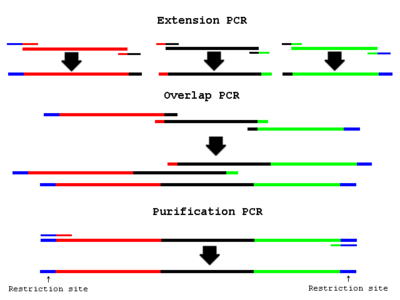
Overview
Create long DNA fragments from shorter ones. This method is also called “Splicing by Overlap Extension” or SOEing.
Procedure
- Design Primers:
- These primers are like bridges between the two parts you want to assemble together.
- You will order two primers which are complements of one another.
- These primers will each have a 60°C Tm with one part and a 60°C Tm with the other part.
- The “end primers” will not have any complements and will likely only have restriction sites.
- “Extension PCR” PCR amplify the necessary fragments separately
- Use a proofreading polymerase enzyme.
- Use an annealing temp of 60°C.
- Clean up the product using a DNA column.
- “Overlap PCR” Use cleaned up fragments as template in a PCR reaction:
- About 1/2 to 3/4 volume of the Overlap PCR reaction should be equimolar amounts of purified fragments.
- Do not use Phusion polymerase. Try Pfu Turbo.
- Do not add any primers; the templates will prime each-other.
- Run 15 PCR cycles without primers.
- Use an annealing temp of 60°C.
- “Purification PCR” Add end primers to the Overlap PCR reaction:
- Continue cycling for another 15-20 rounds.
- Use an annealing temp of 72°C
- Gel extract the correct size fragment.
- Clone into the desired vector.
- Digest
- Ligate
- Transform
- Select
- Sequence